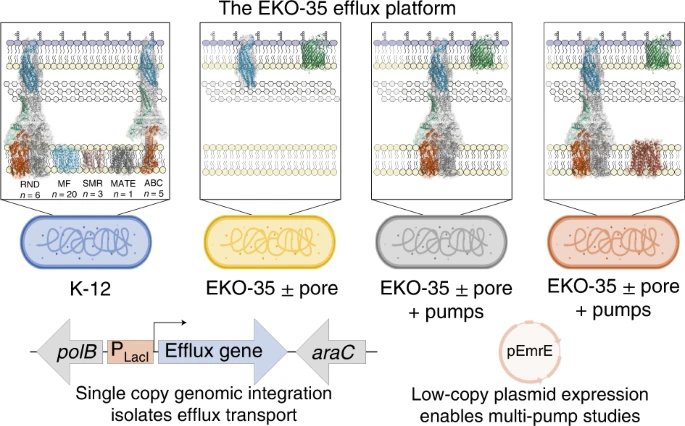
Bacterial Efflux Systems.
Antibiotic resistance is a global health threat. To address this crisis, we need to develop new antimicrobial therapeutics and we need to gain a more thorough understanding of antibiotic resistance mechanisms. Efflux pumps pose a significant challenge for the development of new antibacterial agents. These membrane-spanning proteins enable bacteria to resist the inhibitory action of most clinically used classes of antibiotics. While some efflux pumps are well-characterized, bacterial species harbour large networks of additional and often poorly characterized drug efflux pumps. The complexities of these drug efflux systems preclude studies to elucidate efflux pump functions.
To address these limitations, we are developing tools and platforms to facilitate the study of bacterial drug efflux pumps. Using these tools, we are investigating the physicochemical substrate specificities and physiological functions of these ancient resistance elements. We’re mainly studying Escherichia coli efflux pumps, but we’re expanding this work to include efflux pumps from other organisms too. We will use this information to inform drug discovery endeavours to identify efflux inhibitors and new antibiotics that evade efflux.
In addition, we are curious to explore why these proteins are highly conserved despite numerous pumps being poorly characterized. Indeed, we recently showed that ~80% of E. coli drug efflux pumps are conserved across the species!
The major bacterial drug efflux protein families.